Introduction
On April 12, 2002, the FDA had alerted health care professional about the risk of Enterobacter sakazakii (Cronobacter spp.) infection in hospitalized newborn infants, particularly premature infants or other immuno-compromised infants who fed powdered infant formulas [1–5]. This alert issued by FDA was targeted to concerns for immnuo-compromised infants in hospital settings [1,3–5]. The conventional methods for isolating and identificating E. sakazakii (Cronobacter spp.) from infant formula required enrichment culture for about 48 h, and then subculture to selective agar followed by phenotypic identification [1–3]. Namely, these procedures took up at least 5 days to obtain a result [1–3]. Therefore, in order to isolate and identify E. sakazakii (Cronobacter spp.) rapidly, various studies have been conducted, and various attempts to inhibit the growth of E. sakazakii (Cronobacter spp.) had also been conducted [1,5–7]. Fortunately, Real-Time PCR could provide the rapid-accurate-quantitative analysis for confirming various food-borne pathogens [8–10]. The format was developed for detecting E. sakazakii (Cronobacter spp.) in dried infant formula using by applying the fluorogenic 5' nuclease assay (TaqMan®) with ABI Prism 7000 [10]. Seo and Brackett [10] firstly made the format of probe and primers based on the partial macromolecular synthesis operon of E. sakazakii (Cronobacter spp.) such as the rpsU gene 3' end and the primase (dnaG) gene 5' end [10]. Also, in the Real-Time PCR instrument, the fluorescence measurement is performed in real time and automatically analyzed by the SDS software installed in the sequence detector [8–10]. Therefore, using a Real-Time PCR instrument, results could be quickly confirmed within 2 hours [8–10].
Also, various chromogenic agars have been developed to detect E. sakazakii (Cronobacter spp.). To date, various companies have developed chromogenic agar products for detection of E. sakazakii (Cronobacter spp.), but it is urgently needed to develop a method to solve the problem of false positives and false negatives [11–13]. Among the various chromogenic agars, Druggan-Forsythe-Iversen (DFI, UK) was first developed for the purpose of detecting E. sakazakii (Cronobacter spp.), and is still widely used.
Hence, this study developed and compared Real-Time PCR assays based on the macromolecular synthesis (MMS) gene, for the detection of E. sakazakii (Cronobacter spp.) using the ABI Prism 7000 SDS (Applied Biosystems, USA) instrument with a selective chromogenic agar within 24 hours. And also real-time PCR approaches were compared with the conventional culture method for the detection of E. sakazakii (Cronobacter spp.) in dried infant formula.
Materials and Methods
All strains (E. sakazakii [Cronobacter spp.] and non-E. sakazakii [non-Cronobacter spp.]) were provided by Division of Microbiology, CFSAN, FDA, and these bacterial strains were cultured in tryptic soy broth (TSB; Becton Dickinson, USA) at 37℃. To obtain an initial low cell number, 10-fold serial dilutions were performed in phosphate-buffered saline (PBS, pH 7.2). Viable cell numbers of these strains were determined by direct plating to DFI (Chromogenic E. sakazakii [Cronobacter spp.] agar, CM1055, Oxoid) following incubation at 37℃ for 24 hours. Also, these strains were examined for its response to stress condition and recovery on selective media as DFI.
Various types of dried infant formula were purchased from a grocery store in College Park, USA. Dried infant formula were artificially inoculated at several different target levels, and the inoculated test portions were submitted to the one-enrichment method at Fig. 1, using by DFI as selective media and real-time PCR. Assays were repeated 3 times for all contamination levels including for the blank (negative control). Detection limit was assessed for E. sakazakii (Cronobacter spp.) N6 strain. This stain was selected to present the typical yellow pigmentation on TSA, the green color on DFI, and positive on Real-Time PCR.
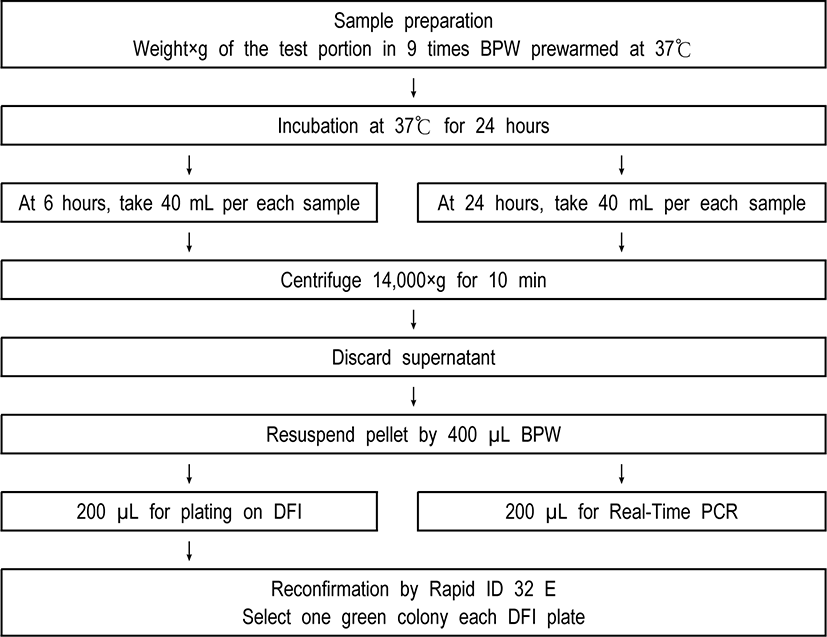
A real-time PCR assay developed by Seo and Brackett [10] targeting the dnaG gene on the MMS operon of E. sakazakii (Cronobacter spp.) was modified and used to confirm identification of each sample. Two sets of each sample (200 µL) from one-time enrichment broth were centrifuged at 14,000×g for 10 min.
Negative control (filter sterilized water) and positive control (1 mL of E. sakazakii (Cronobacter spp.) with over 108 CFU/mL) were included in every assay. The pellets of cell were resuspended in PreMan Ultra (Applied Biosystems) and then placed in the boiling water (about 100℃) during ten min. Hence, samples were cooled for 2 min at ambient temperature and also centrifuged at 14,000×g for 10 min again.
Forty-five microliter of PCR mix was combined by 5 µL of forward primer, 5 µL of reverse primer, 5 µL of TaqMan probe, 5 µL of filter sterilized water, and 25 µL TaqMan Universal PCR Master Mix in a 96-well microwell plate, respectively. And then, 5 µL of the supernatant fluids was transferred to the prepared PCR mix. Next, the 96-well microwell plates were sealed with optical adhesive covers and were analyzed by ABI Prism 7000 SDS (Applied Biosystems). The reaction was run at 50℃ for 2 min and then 95℃ for 10 min, followed by 40 cycles of 95℃ for 15 s and 60℃ for 60 s.
Furthermore, it was also investigated whether the ABI Prism 7000 SDS format designed for detecting E. sakazakii (Cronobacter spp.) was applicable to real-time PCR devices such as rotor gene, smart cycler, and light cycler produced by other companies.
Also, the presumptive colony of E. sakazakii (Cronobacter spp.) shown green color was taken from each DFI agar plate and then were biochemically identified by the Rapid ID 32 E strips (bioMérieux), which were incubated at 37℃ for approximately 4 hours.
Varied cell numbers of E. sakazakii (Cronobacter spp.) along with defined numbers of either Salmonella Enteritidis or Enterobacter cloacae were spiked simultaneously into×(ex. 25)g of dried infant formula in 9 times (ex. 25×9 times) mL buffered peptone water (BPW), and then followed by one-time enrichment method (Fig. 1).
Results & Disscussion
The purpose of this study was to detect E. sakazakii (Cronobacter spp.) within 24 hours. The first is the result of a comparative experiment on the most suitable enrichment broth for E. sakazakii (Cronobacter spp.). First, mLST, universal pre-enrichment broth, water including 0.85% NaCl, buffered peptone water, widely used as an enrichment medium for detecting microorganisms, and R&F enrichment broth sold as a selective enrichment medium for E. sakazakii (Cronobacter spp.) were compared respectively (Table 1).
| Incubation time | Type of enrichment media | |||||
|---|---|---|---|---|---|---|
| M | R | U | W | B | Colony color on DFI | |
| 0 hour | - | - | - | - | - | - |
| 6 hours | - | 2.65 | 2.14 | 3.1 | 3.4 | Green1) |
| 18 hours | 6.7 | 9.04 | 9.41 | 9.47 | 9.59 | Green |
Growth of E. sakazakii (Cronobacter spp.) was confirmed through DFI, an E. sakazakii (Cronobacter spp.) selective chromogenic agar, while culturing for 0 hours, 6 hours, and 18 hours. mLST, R&F enrichment broth, universal pre-enrichment broth, water including 0.85% NaCl, and buffered peptone water were all detected at 18 hours, and only R&F enrichment broth, universal pre-enrichment broth, water including 0.85% NaCl, and buffered peptone water were detected at 6 hours. As a result, buffered peptone water showed the best results (Table 1).
In addition, an experiment was conducted in which vancomycin, an antibiotic capable of inhibiting Gram-positive bacteria, was added to various enrichment broth. As a result, there was no statistically significant difference between the absence of vancomycin and the addition of vancomycin (final concentration is 8 µg/mL) (data not shown).
Next, the inclusivity and exclusivity were compared using various methods used to identify E. sakazakii (Cronobacter spp.) (Table 2). Summarizing the results obtained in this study, nutrient agar did not show any differentiation because all microorganisms were able to grow. However, since DFI is a chromogenic agar that can detect E. sakazakii (Cronobacter spp.), it was able to specifically observe green colony. Also, Ripid ID 32E was able to distinguish E. sakazakii (Cronobacter spp.). Furthermore, real-time PCR was able to confirm the presence of E. sakazakii (Cronobacter spp.) in a short time (Table 2).
Therefore, to summarize the results of this experiment, the optimal combination for detecting E. sakazakii (Cronobacter spp.) is to check whether positive or negative by the color of the colony with DFI agar, and to check whether it is E. sakazakii (Cronobacter spp.) or not by using real-time PCR at the same time. Then, if necessary, additional confirmation of green colonies on DFI agar by Rapid ID 32E method is considered to be able to accurately identify E. sakazakii (Cronobacter spp.).
In order to detect E. sakazakii (Cronobacter spp.) within 24 hours, the method of determining whether positive or negative for E. sakazakii (Cronobacter spp.) is positive or negative with various newly developed devices such as real-time PCR in the enrichment medium for 6 hours after starting the enrichment culture broth is considered very important. In particular, securing a large number of bacteria is the biggest challenge in order to confirm positive or negative of E. sakazakii (Cronobacter spp.) in the enriched culture broth for 6 hours. Therefore, the centrifugation technique, which is a method commonly used in Escherichia coli testing, was applied to the detection of E. sakazakii (Cronobacter spp.) [14]. Therefore, the enrichment culture was started for the samples to which various concentrations of E. sakazakii (Cronobacter spp.) were added, and the enriched culture broth was centrifuged at 6 hours and 24 hours, respectively, and those without centrifugation were compared with each other (Table 3).
The method of centrifuging the enriched culture broth for both 6 and 24 hours showed a good result, which was very helpful in determining E. sakazakii (Cronobacter spp.), as the number of bacteria was detected with a statistically significant difference (p<0.05).
Next, in order to secure more bacterial counts of E. sakazakii (Cronobacter spp.), we compared the results of bacterial counts in DIF, a selective chromogenic agar, and real-time PCR for E. sakazakii (Cronobacter spp.), when the centrifuged amount of the enriched culture medium was expanded to 40 mL (Table 4). As a result, as the amount of centrifugation of the enrichment culture increased, better results were obtained (Table 4). In addition, according to previous study, the real-time PCR results were compared when the centrifuged amounts of the enriched culture medium were 1 mL, 10 mL, and 40 mL at 6 hours and 24 hours, respectively. As expected, the best results were obtained when the amount of centrifugation was 40 mL (Data not shown).
The real-time PCR used in this study was manufactured based on the ABI Prism 7000 SDS. Therefore, we tested whether this format of real-time PCR, which was made to identify E. sakazakii (Cronobacter spp.), can be applied to other real-time PCR devices (Table 5). This is because each laboratory has real-time PCR devices produced by various companies.
A total of four real-time PCR devices compared in this study are ABI Prism 7000 SDS, rotor gene, smart cycler, and light cycler. As shown in Table 5, the format of ABI Prism 7000 SDS, which can discriminate positive and negative for E. sakazakii (Cronobacter spp.), was directly applied to rotor gene and smart cycler to accurately discriminate positive and negative. However, it was shown that it was not applied in the case of light cycler (Table 5).
In conclusion, this study established a method using a selective chromogenic agar and real-time PCR simultaneously to detect E. sakazakii (Cronobacter spp.) within 24 hours. An enrichment culture broth capable of selectively enriching E. sakazakii (Cronobacter spp.) was investigated. In addition, by applying the centrifugation method, E. sakazakii (Cronobacter spp.) was detected even if the incubation time was 6 hours. In order to further improve this method, it is necessary to set the number of selective chromogenic agar to at least two. Additional rapid & accurate detection methods should also be developed. In addition, further studies are needed to increase the sensitivity of real-time PCR for confirming E. sakazakii (Cronobacter spp.). Furthermore, it is necessary to actively use upgraded biochemical methods (for example, Matrix-Assisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometer, etc) that are currently being developed in various ways [15]. Further research is needed so that the method developed in this study can be used in the detection of other food-borne poisoning bacteria.






