Introduction
Foodborne diseases attributed to bacterial pathogens such as Bacillus cereus, Clostridium perfringens, Staphylococcus aureus and Escherichia coli continue to pose considerable challenges to global public health systems [1–6]. Despite improvements in sanitation, preservation, and food processing technologies, these organisms are frequently implicated in contamination events, particularly in ready-to-eat (RTE) or minimally processed food products [7,8]. Even at low contamination levels, their presence can undermine food safety, trigger product recalls, and cause reputational and financial harm to producers [9].
To mitigate these risks, regulatory agencies—including those in South Korea—have increasingly integrated quantitative microbial risk assessment (QMRA) frameworks into food safety control strategies [10–13]. A critical aspect of such systems involves the reliable enumeration of pathogenic microorganisms using selective media that can suppress non-target flora while facilitating accurate quantification of the organism of interest [14–17].
Currently, the Korean Food Code recommends microbial detection protocols largely derived from international frameworks, including the U.S. FDA’s Bacteriological Analytical Manual (BAM), International Organization for Standardization (ISO) guidelines, and AOAC International methods [18–20]. However, these protocols were often optimized for Western-style food matrices and may not be ideally suited to the complex compositions of Korean foods, which include fermented, seasoned, and plant-based products. Additionally, variations in the formulation of selective agents, incubation conditions, and colony morphology across different media can significantly impact the accuracy and reproducibility of microbial detection.
To overcome these limitations, this study was designed to evaluate the quantitative recovery performance of multiple selective media under harmonized conditions using BioBall™, a precision-manufactured inoculum containing verified colony-forming unit (CFU) counts of ISO- and AOAC-certified bacterial reference strains. By eliminating inoculum variability and standardizing experimental parameters, the objective was to generate robust, reproducible data that can support future revisions to national microbiological testing standards and potentially expand the pool of officially accepted media.
Materials and Methods
To ensure uniform inoculation across test conditions, BioBall™ (BioMérieux, France), an ISO- and AOAC-certified microbial reference standard, was employed. Each BioBall™ unit contained precisely 30 viable colony-forming units (CFU) of a defined bacterial strain. Reference strains were sourced from internationally recognized culture collections, including the American Type Culture Collection (ATCC) and the National Collection of Type Cultures (NCTC). Prior to experimental use, the viability and cell count of each BioBall™ lot were verified via plating on non-selective media (Table 1).
Each unit was rehydrated in 0.1 mL of sterile distilled water and thoroughly mixed by gentle vortexing to achieve homogeneous cell suspension. A 0.1 mL aliquot of this suspension was then aseptically spread onto the surface of the selective media using a sterile glass spreader, resulting in an expected inoculum of approximately 30 CFU per plate.
Ten selective media were evaluated for their ability to support the growth of four major foodborne pathogens (Table 2).
All media were chosen based on their regulatory relevance or widespread use in industry. Chromogenic and dry film formats were included to evaluate modern alternatives with simplified interpretation and procedural benefits.
After inoculation, the plates were incubated under organism-specific conditions optimized for growth and selectivity. Incubation temperatures and atmospheres were as follows: Bacillus cereus is a facultative anaerobic, Gram-positive rod-shaped bacterium. It is typically incubated under aerobic conditions at 30°C for 24 to 48 hours. B. cereus forms spores and is known for producing emetic and diarrheal toxins, making it an important pathogen in food safety monitoring. C. perfringens is an obligate anaerobic, Gram-positive rod that requires strict anaerobic conditions at 37°C for 24 hours for effective growth. Because it is highly sensitive to oxygen, specialized anaerobic chambers or jars are used during incubation. It is a common cause of foodborne illness, particularly in meat and poultry products. Staphylococcus aureus is a Gram-positive coccus that grows well under aerobic conditions at 37°C for 24 to 48 hours. It is capable of producing heat-stable enterotoxins that can cause food poisoning and is widely monitored in food hygiene testing. And Escherichia coli is a facultative anaerobic, Gram-negative rod commonly used as an indicator of fecal contamination. It is incubated under aerobic conditions at 35°C to 37°C for 24 hours. E. coli grows rapidly and is routinely tested in both food and water microbiological assessments.
Following incubation, colony-forming units were enumerated visually. Recovery rates were calculated as the mean CFU observed per plate in comparison with the expected input of 30 CFU. Each condition was tested in five biological replicates to ensure statistical robustness.
To evaluate differences in recovery efficiency among selective media for each pathogen, ANOVA was performed. Where significant differences were identified (p<0.05), Tukey’s post hoc test was applied to determine pairwise significance. All statistical analyses were conducted using SPSS software version 18.0 (IBM, USA), and significance was set at p<0.05.
Results and Discussion
Three selective media—mannitol-yolk-polymyxin B agar (MYP), polymyxin egg yolk mannitol bromothymol blue agar (PEMBA), and a commercially available chromogenic medium—were assessed for their efficacy in recovering Bacillus cereus from a standardized BioBall™ inoculum. All media demonstrated high recovery rates, exceeding 80% based on the 30 CFU reference input. Among them, PEMBA exhibited the highest mean recovery (125 CFU), followed closely by MYP and the chromogenic medium, both yielding an average of 123 CFU (Fig. 1).
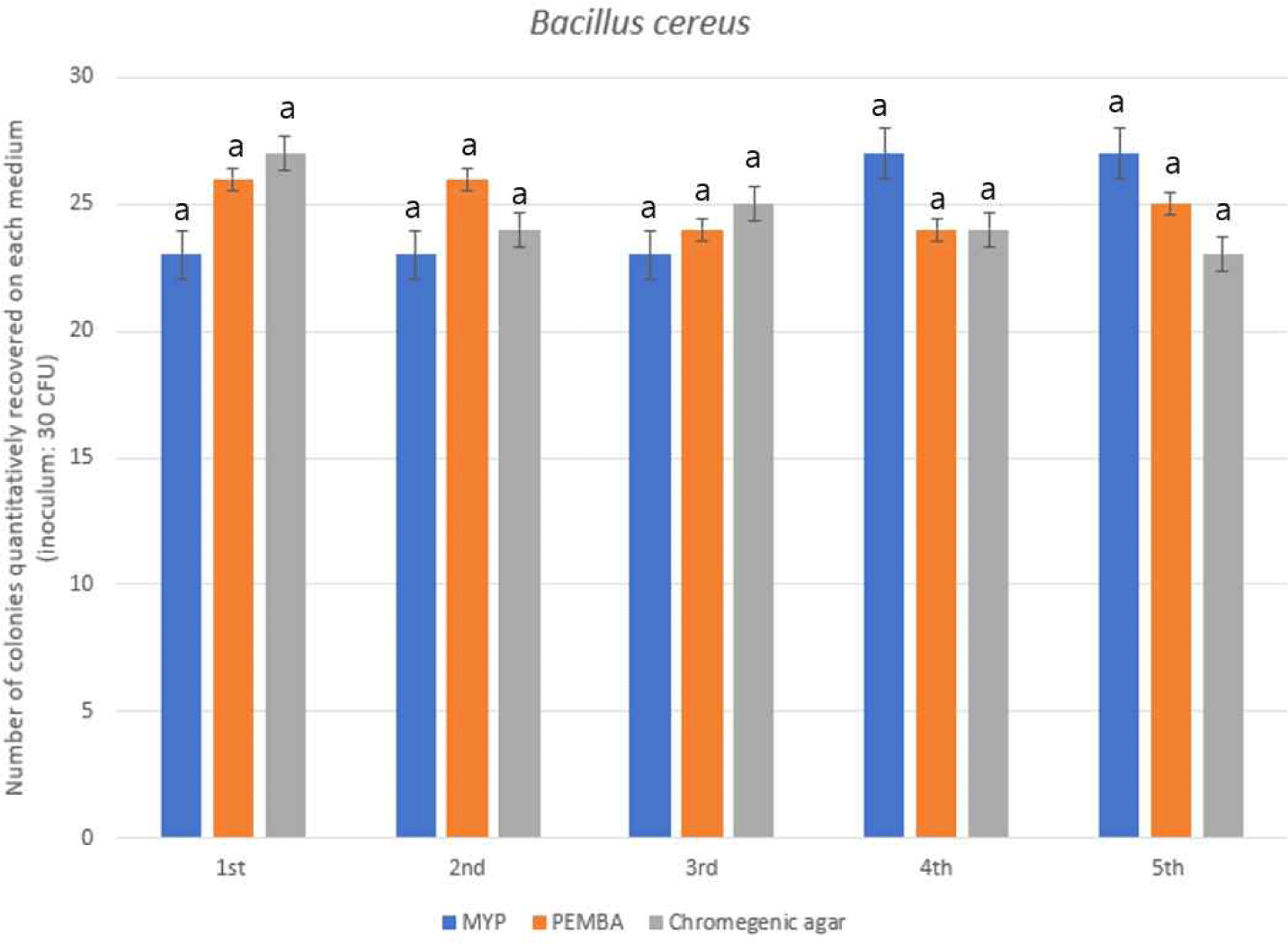
Despite these numerical variations, statistical analysis via one-way ANOVA indicated no significant difference in recovery performance among the tested media (p>0.05). This suggests that all three formulations offer comparable efficiency in enumerating B. cereus under controlled conditions.
MYP, the conventional medium cited in the Korean Food Code, remains widely utilized due to its ability to differentiate B. cereus through egg-yolk lecithinase reaction and mannitol fermentation. PEMBA, developed to improve visual contrast and colony discrimination, may offer practical advantages in routine testing, such as enhanced readability and lower background interference. The chromogenic medium, though relatively novel, matched the performance of its traditional counterparts and offered the additional benefit of colorimetric identification—an asset for rapid screening and reduced analytical subjectivity.
Collectively, these findings support the interchangeable use of all three media for B. cereus enumeration, providing flexibility in both regulatory and industrial testing frameworks depending on laboratory resources and analytical objectives.
To assess the recovery of C. perfringens, three media were compared: Tryptose Sulfite Cycloserine (TSC) agar, Shahidi-Ferguson Perfringens (SFP) agar, and a commercial chromogenic medium. TSC yielded the highest recovery rate at 96%, surpassing SFP (84%) and chromogenic medium (71%), as shown in Fig. 2. Statistical analysis confirmed a significant difference between TSC and the chromogenic medium (p<0.05), suggesting reduced effectiveness of the latter under the tested conditions.
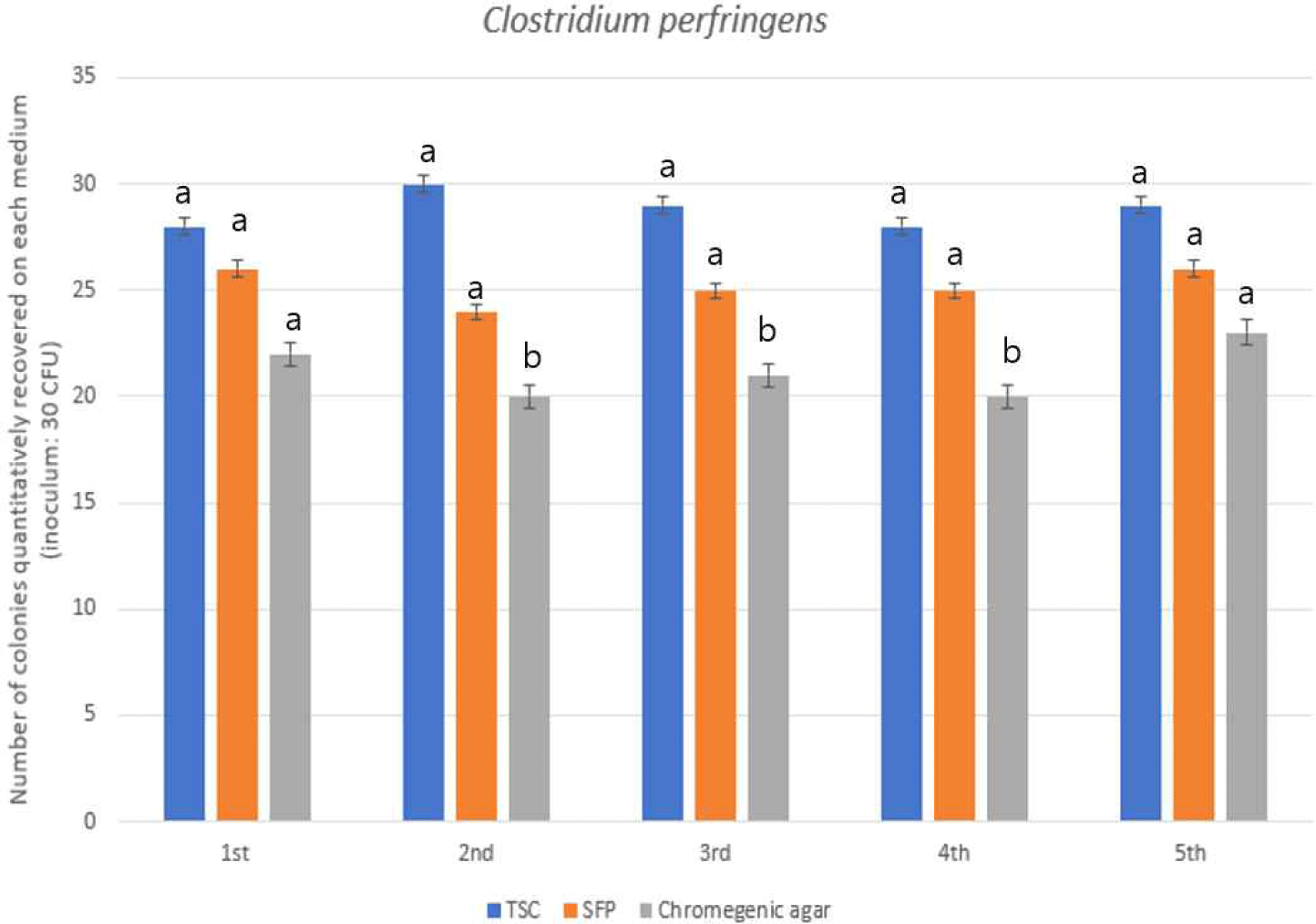
TSC agar is widely considered the reference medium for C. perfringens due to its selective agents—cycloserine to inhibit competing anaerobes and sulfite/iron components that produce a distinctive black precipitate with the target organism. SFP, although similar in design, exhibited slightly diminished performance, potentially due to lower inhibitory strength.
The chromogenic medium, while offering visual convenience through enzyme-specific color changes, demonstrated inferior recovery. This may stem from limitations in supporting strict anaerobic growth or lack of key reductive substrates.
These results reinforce the utility of TSC for sensitive and accurate detection of C. perfringens. While SFP may serve as a practical secondary option in standard testing workflows, caution should be exercised with chromogenic media, particularly in samples with complex or competitive background flora.
Four media were evaluated for their ability to support the quantitative recovery of Staphylococcus aureus: Mannitol salt agar (MSA), Baird-Parker Agar (BP), baird-parker with rabbit plasma fibrinogen (BP-RPF), and a chromogenic formulation specific to S. aureus. All tested media demonstrated comparable performance, with recovery rates ranging from 89.3% to 93.3%, and no statistically significant differences were observed (p>0.05; Fig. 3).
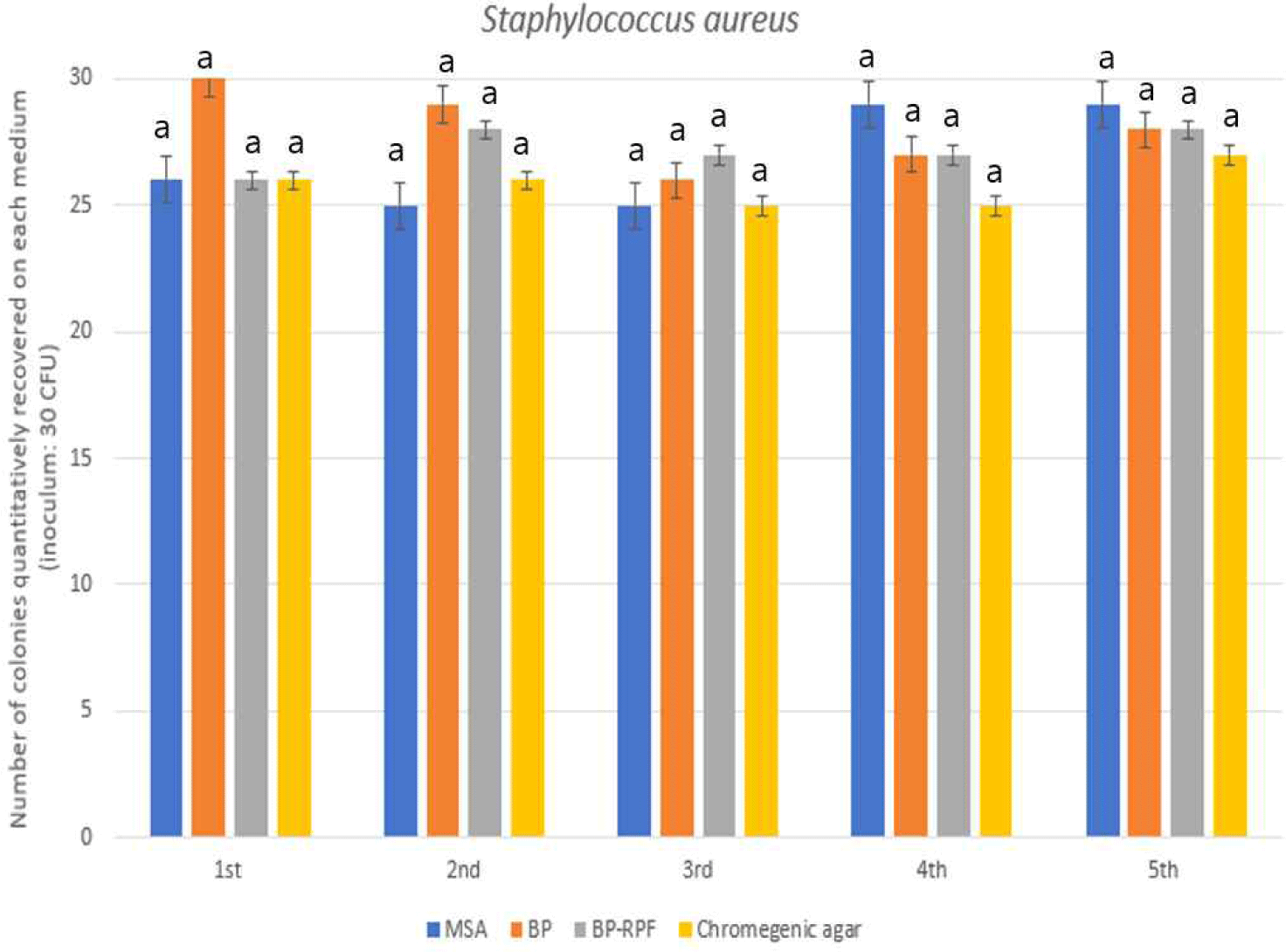
Each medium leverages a unique physiological or biochemical trait of S. aureus to achieve selectivity. MSA employs high salt concentrations and mannitol fermentation as differentiation mechanisms. BP and its RPF-supplemented variant utilize tellurite reduction and lecithinase activity, with the fibrinogen supplement further enhancing detection of coagulase-positive strains. Chromogenic media are designed to visually identify S. aureus through substrate-specific color development, streamlining colony identification.
The consistent results across all media affirm their applicability for routine enumeration of S. aureus. Selection of the most appropriate medium may thus be driven by operational considerations—such as analysis speed, visual clarity, or the need for confirmatory testing—rather than by performance differentials.
To evaluate the performance of selective media for Escherichia coli, four approaches were examined: a chromogenic medium targeting β-glucuronidase activity, eosin methylene blue (EMB) agar, MacConkey agar, and a commercially available dry film platform. All methods yielded high recovery rates between 89.3% and 93.3%, with no significant differences observed among the groups (p>0.05; Fig. 4).
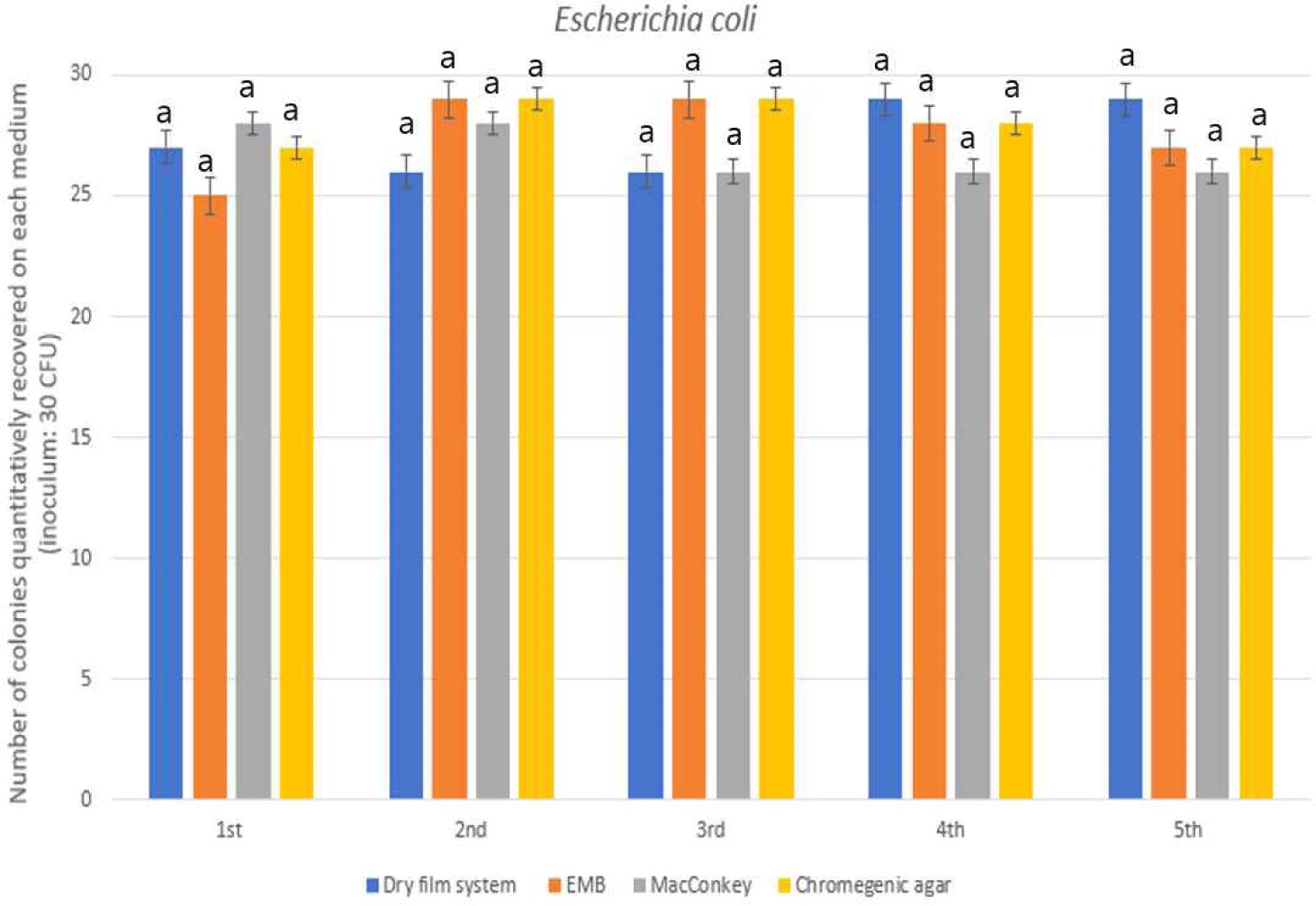
Chromogenic media typically detect E. coli via hydrolysis of β-glucuronide substrates, resulting in colored colonies that enable rapid, direct identification. EMB and MacConkey agars are well-established traditional media that utilize lactose fermentation and pH indicators to differentiate E. coli from non-coliforms. The dry film method, based on enzyme-substrate reactions embedded in a dehydrated matrix, provided equally reliable results with the added advantage of simplified handling.
The uniformity in performance suggests that all evaluated methods are viable for the enumeration of E. coli in food matrices. While conventional agars remain cost-effective and robust, chromogenic and dry film technologies offer enhanced visual clarity and operational efficiency, particularly in laboratories with limited resources or high-throughput demands.
This comprehensive evaluation across four major foodborne pathogens—Bacillus cereus, C. perfringens, Staphylococcus aureus, and Escherichia coli—revealed that a wide range of selective media, including both conventional formulations and newly developed alternatives, exhibited satisfactory performance in terms of recovery efficiency. Notably, several non-regulatory media demonstrated recovery rates that were comparable to, or in some cases exceeded, those of the selective media currently endorsed by the Korean Food Code. These results highlight the practical feasibility of incorporating non-official media into quantitative food microbiology workflows.
Among the media evaluated, chromogenic formulations showed consistently high efficacy across all tested pathogens. Designed with chromogenic substrates that produce organism-specific coloration, these media offered distinct advantages in colony differentiation and visual discrimination between target organisms and background microflora [21–23]. This feature not only improved accuracy but also reduced the necessity for confirmatory identification procedures, thereby minimizing potential misidentification errors and expediting microbial enumeration processes [22,23]. As such, chromogenic media represent a significant advancement in the streamlining and automation of routine microbiological testing [24]. Dry film media, including Petrifilm™ and similar rapid detection platforms, also exhibited excellent recovery rates and high reproducibility across replicates [25]. Although these formats deviate from traditional agar-based methodologies, they offer practical advantages, particularly in high-throughput or resource-limited settings. Their compactness, ease of storage, and minimal preparation requirements contribute to reduced labor, lower procedural variability, and faster processing times. The consistent performance observed in this study supports their integration into standardized testing protocols, especially in industrial environments where efficiency is paramount.
A key methodological strength of this study was the utilization of BioBall™, a commercially available microbial reference standard certified by both ISO and AOAC [26]. This product ensures consistent inoculum levels across all replicates, thereby eliminating variability associated with manual inoculation. By standardizing the viable cell counts used in testing, BioBall™ significantly enhanced the reproducibility and reliability of experimental outcomes, allowing for more objective and precise assessment of media performance [26]. The use of such standardized reference materials sets a benchmark for future comparative studies and contributes to methodological rigor in food microbiological validation.
Collectively, these findings suggest that several alternative selective media—particularly chromogenic and dry film formats—exhibit sufficient sensitivity, selectivity, and repeatability to serve as viable substitutes for currently recognized regulatory media [25–27]. The combination of technical reliability and operational efficiency makes these alternatives not only scientifically sound but also cost-effective and logistically favorable for routine implementation in food safety monitoring [23,28,29].
From a regulatory perspective, these results underscore the need to transition toward more flexible, evidence-based validation frameworks. As food production systems evolve and new microbial hazards emerge, reliance on a fixed list of prescribed media may no longer suffice. Updating the official compendium of approved media to reflect performance-driven evaluations can enhance adaptability and responsiveness to emerging challenges. This approach would support the modernization of food safety standards and encourage innovation within the diagnostic microbiology sector.
Incorporating scientifically validated alternative media into official testing protocols would benefit both regulatory agencies and industry stakeholders by improving analytical efficiency and data reliability. Moreover, such initiatives would align with international efforts toward harmonization of food microbiological methods, thereby facilitating cross-border recognition of test results and promoting global food trade.
Conclusion
This study presented a detailed comparative assessment of selective media used to quantify four key foodborne pathogens: Bacillus cereus, C. perfringens, Staphylococcus aureus, and Escherichia coli. By incorporating BioBall™—a certified, fixed-count microbial reference standard—into all assays, the experiments ensured reproducibility and minimized variation across trials.
The results showed that several non-official media performed as well as, or in some cases better than, those currently endorsed by the Korean Food Code. For B. cereus, MYP, PEMBA, and chromogenic media demonstrated equivalent efficacy. In the case of C. perfringens, TSC achieved the highest recovery, followed by SFP, while the chromogenic medium lagged behind. For S. aureus, all tested media—including MSA, BP, BP-RPF, and chromogenic formats—yielded consistent results. Finally, for E. coli, traditional agars and dry film formats performed comparably, with chromogenic methods offering additional interpretive convenience.
Moreover, the operational advantages of chromogenic and dry film technologies—such as faster turnaround, reduced analytical subjectivity, and lower training demands—highlight their applicability in modern testing laboratories (Table 3). The use of standardized inoculation such as BioBall™ further enhances the reliability of comparative media studies and supports harmonized validation protocols.
Altogether, the findings suggest that expanding the roster of officially approved selective media—particularly to include validated chromogenic and dry film formats—could improve the accuracy, flexibility, and efficiency of microbial testing in regulatory and industrial settings. These insights could inform future revisions to microbiological testing standards in Korea and contribute to broader global harmonization efforts.
